3D Cell Cultures
Nanolive imaging requires samples to be transparent in order for the light to pass through and be collected underneath; thus it is normally suited for high spatio-temporal resolution imaging of live 2D cell cultures. However, this rule of thumb has some exceptions that we will analyse on this page.
3D cell culture systems provide the natural microenvironment of cells, which leads to the obtention of data that is physiologically relevant and more predictive for in vivo tests.
3D cell cultures allow for an improved visualization of cell-cell and cell-matrix interactions. Cell structure and cell populations in 3D cell cultures represent a closer approach to in vivo architecture than traditional 2D monolayer cultures.
Some fields of applications such as drug discovery, stem cell research and cancer cell biology among others are strongly susceptible to benefit from the advantages of 3D cell culture systems.
Mesenchymal stem cell interacting with 3D hydrogel
For this video, mesenchymal stem cells were grown in 3D PEG hydrogels of varying degrees of stiffness, which were kindly provided by Dr. Sonja Giger and Prof. Matthias Lutolf from EPFL, Switzerland. The cells were then imaged for 48 hours at an acquisition rate of one image every 30 seconds. A post-processing colour scale was applied to data in the z-axis, to add spatial information to our images. Blue indicates where the cell is closer to the surface of the gel, while pink indicates where it is further away.
The solid gel had no impact on image quality; cells spread further and were more mobile on soft gels than stiff gels but were equally healthy on both. Images taken of cells on one of the soft hydrogels were particularly interesting. The surface of the gel in question had slight indentations on the surface creating a more complex nanotopography. The cell can be observed pulling and pushing on gel pieces and occupying the holes on the surface (we marked these moments in the video with circles). These observations show that the combination of 3D hydrogels and holotomography can open the door for novel observations to be made about how stem cells sense and interact with their physical environment.
HeLa cells in gel
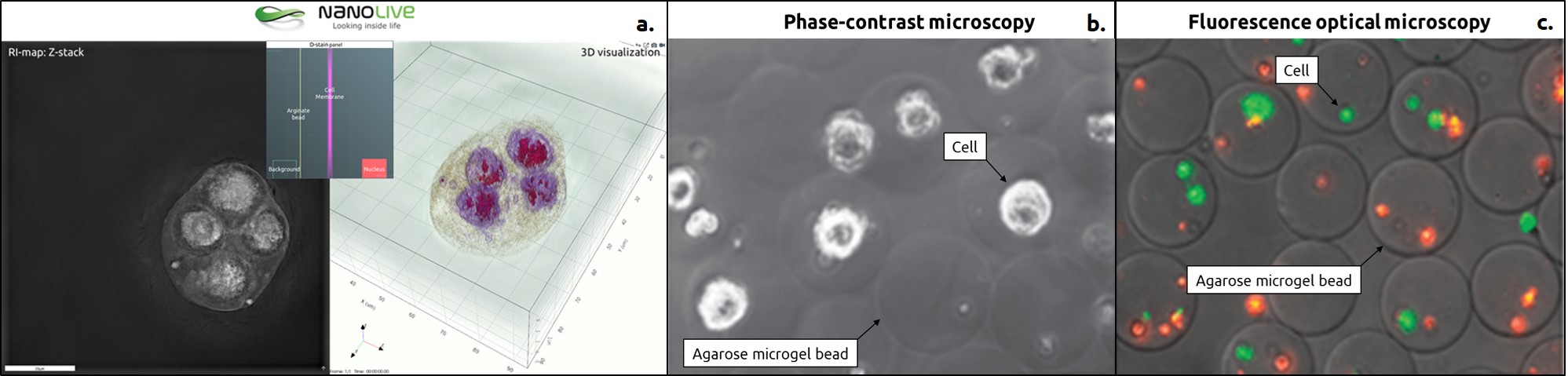
Image a shows HeLa cells encapsulated in alginate beads suspended in DMEM solution and visualized through a glass coverslip. The alginate beads were generated using sciDROP PICO technology mounted on a sciFLEXARRAYER S3 (SCIENION AG, Germany). Image b. & c. show 100 μm-diameter agarose microgels encapsulating mESs (mouse embryonic stem cells). High-throughput combinatorial cell co-culture using microfluidics, 28 Apr 2011, Ethan Tumarkin et al.
3D alginate beads with E.coli
Nanolive’s 3D Cell Explorer enables accurate and quantitative 4D spatio-temporal monitoring of bacteria cultures. It also allows to monitor several E.coli bacteria colonies growing into 3D systems stain-free (e.g. alginate beads); monitor the volume of a single bacterium or of a growing bacteria colony and image and discriminate multi-layers of bacteria totally stain-free.
This video shows E.coli bacteria embedded in alginate beads (generated by Encapsulation Unit – Var J30, 40-70 microns diameter). The beads were mounted on a slide into Minimum Media (MM) diluted 1:4 with distilled water (plus 1% v/v Fumarate 100mM) . The time-lapse imaging experiment was conducted with a standard top-stage incubator set to 37°C and 90% humidity for 3 hours, capturing images every 10 minutes.
Image courtesy of prof. Van Der Meer Jan Roelof (Département de microbiologie fondamentale, UNIL, Lausanne, Switzerland).
Micro-pillar: Cells on-a-chip
The 3D Cell Explorer enables the analysis of morphological changes and membrane remodeling due to functionalized surfaces.
This video lets you observe how the adhesion of the cell to the substrate is guided by the nanostructures on the device surfaces.
The cell-on-a-chip technology can provide micro-scale complex structures and well-controlled parameters to mimic the in vivo environment of cells. The 3D Cell Explorer offers the great potential of getting a non invasive, 3D real time imaging of cells directly on this glass chips.
Collagen matrices
Collagen is the main protein present in the extracellular matrix. 3D cell cultures in collagen matrix mimic in vivo environment. Cell-matrix interactions in 3D cell cultures represent a closer approach to in vivo cell behaviour. The 3D Cell Explorer allows for high spatio-temporal resolution images of 3D cell cultures over time.
Dendritic cells in Matrigel
This video demonstrates 3D visualization of Inactive and Activated dendritic cells in 3D matrix (Matrigel).
Video Library
Multilayer E.coli bacteria
